-Search query
-Search result
Showing 1 - 50 of 116 items for (author: nans & a)
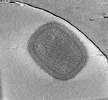
EMDB-18916: 
Cryotomogram of mature Vaccinia virus (WR) virion
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18917: 
Subtomogram average of the Vaccinia virus (WR) portal complex in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18918: 
Subtomogram average of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8r5i: 
In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15596: 
In situ subtomogram average of Vaccinia virus (WR) palisade, all virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15597: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from CEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15598: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IMVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15599: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15600: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (immature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15601: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15602: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8arh: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-26862: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Method: helical / : Li S, Nanson JD, Manik MK, Gu W, Landsberg MJ, Ve T, Kobe B

PDB-7uxu: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Method: helical / : Li S, Nanson JD, Manik MK, Gu W, Landsberg MJ, Ve T, Kobe B

EMDB-13978: 
S. cerevisiae CMGE nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-13988: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-14439: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

PDB-7qhs: 
S. cerevisiae CMGE nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

PDB-7z13: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-13439: 
Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon
Method: single particle / : Kaczmarska Z, Czarnocki-Cieciura M, Rawski M, Nowotny M

EMDB-13440: 
Cryo-EM helical reconstruction of E. coli TnsB in complex with right end fragment of Tn7 transposon
Method: helical / : Czarnocki-Cieciura M, Kaczmarska Z

EMDB-26322: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

PDB-7u32: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

EMDB-26191: 
Cryo-EM structure of human SARM1 TIR domain in complex with 1AD
Method: single particle / : Saikot FK, Kobe B, Ve T, Nanson JD, Gu W, Luo Z, Brillault L, Landsberg MJ

EMDB-14453: 
MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution
Method: single particle / : Ballandras-Colas A, Nans A, Cherepanov P

EMDB-24272: 
Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD)
Method: single particle / : Kerry PS, Nanson JD, Adams S, Cunnea K, Bosanac T, Kobe B, Hughes RO, Ve T

EMDB-24273: 
Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains)
Method: single particle / : Kerry PS, Nanson JD, Adams S, Cunnea K, Bosanac T, Kobe B, Hughes RO, Ve T

EMDB-24274: 
Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (SAM-TIR:1AD)
Method: single particle / : Kerry PS, Adams S, Cunnea K, Brearley A, Bosanac T, Hughes RO, Ve T

PDB-7nak: 
Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD)
Method: single particle / : Kerry PS, Nanson JD, Adams S, Cunnea K, Bosanac T, Kobe B, Hughes RO, Ve T

PDB-7nal: 
Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains)
Method: single particle / : Kerry PS, Nanson JD, Adams S, Cunnea K, Bosanac T, Kobe B, Hughes RO, Ve T

EMDB-13176: 
3.0 A resolution structure of a DNA-loaded MCM double hexamer
Method: single particle / : Greiwe JF, Locke J, Nans A, Costa A

EMDB-13211: 
Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase
Method: single particle / : Greiwe JF, Locke J, Nans A, Costa A

PDB-7p30: 
3.0 A resolution structure of a DNA-loaded MCM double hexamer
Method: single particle / : Greiwe JF, Miller TCR, Martino F, Costa A

PDB-7p5z: 
Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase
Method: single particle / : Greiwe JF, Miller TCR, Martino F, Costa A

PDB-7pel: 
CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma
Method: single particle / : Barski M, Pye VE, Nans A, Cherepanov P, Maertens GN

EMDB-12817: 
Chaetomium thermophilum Chl1 Helicase
Method: single particle / : Hodakova Z, Singleton MR

EMDB-12585: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12586: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12587: 
Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

PDB-7nt9: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

PDB-7nta: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

PDB-7ntc: 
Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-11928: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, Grabe G, Holden D, Nans A, Rosenthal P, Rouse S, Beeby M

PDB-7awa: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, Grabe G, Holden D, Nans A, Rosenthal P, Rouse S, Beeby M

PDB-7beq: 
MicroED structure of the MyD88 TIR domain higher-order assembly
Method: electron crystallography / : Clabbers MTB, Holmes S, Muusse TW, Vajjhala P, Thygesen SJ, Malde AK, Hunter DJB, Croll TI, Nanson JD, Rahaman MH, Aquila A, Hunter MS, Liang M, Yoon CH, Zhao J, Zatsepin NA, Abbey B, Sierecki E, Gambin Y, Darmanin C, Kobe B, Xu H, Ve T

EMDB-23278: 
Cryo-EM structure of ligand-free Human SARM1
Method: single particle / : Nanson JD, Gu W

PDB-7ld0: 
Cryo-EM structure of ligand-free Human SARM1
Method: single particle / : Nanson JD, Gu W, Luo Z, Jia X, Landsberg MJ, Kobe B, Ve T

EMDB-11777: 
RET/GDF15/GFRAL extracellular complex negative stain envelope
Method: single particle / : Adams SE, Earl CP, McDonald NQ

EMDB-11822: 
RET/GDNF/GFRa1 extracellular complex Cryo-EM structure
Method: single particle / : Adams SE, Earl CP, Purkiss AG, McDonald NQ

PDB-7aml: 
RET/GDNF/GFRa1 extracellular complex Cryo-EM structure
Method: single particle / : Adams SE, Earl CP, Purkiss AG, McDonald NQ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
